Integrating EDRR surveillance with eDNA metabarcoding
Mahon, A.R., Grey, E.K. and Jerde, C.L., 2022. Integrating invasive species risk assessment into environmental DNA metabarcoding reference libraries. Ecological Applications, p.e2730. https://doi.org/10.1002/eap.2730
Summary written by Justin Dalaba, edited by Audrey Bowe
Summary
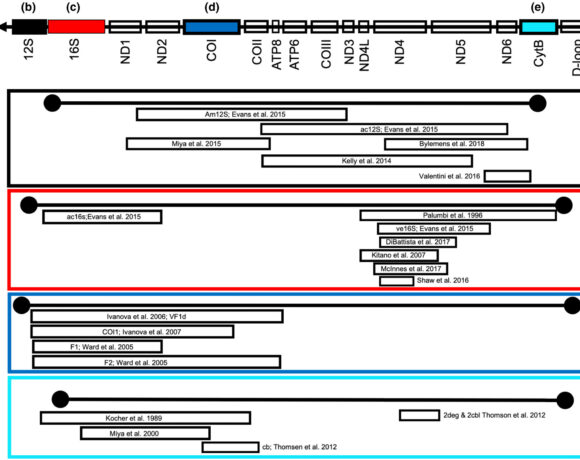
Environmental DNA (eDNA) metabarcoding is a novel method of determining species presence through DNA extracted from water, air, or soil samples. While eDNA use is expanding, there is much still to be developed, including accurate reference sequence libraries to confirm identification and ensure comprehensive coverage for a particular region. For the well-studied Laurentian Great Lakes region, these questions needed addressing: How complete are current reference libraries for eDNA assessments of biodiversity and invasive species? Can we confidently integrate biodiversity monitoring with early detection of aquatic invasive species? In this recent study, Mahon et al. 1) merge two databases (GAPeDNA and ROTH) to provide a comprehensive list of currently known fish species found in the Laurentian Great Lakes region, 2) look at both global and regional risk assessments and evaluate available reference libraries, 3) screen for sequence coverage of 23 common fish species primer pairs, and 4) compare sequences for the three most commonly used target genes of established Great Lakes species in order to quantify species-specificity. They then synthesized this information to find that 30 out of 278 potentially invasive and established species lack reference sequences and should be prioritized to prevent potential false negative or missed detections. Overall, their results demonstrate that even in the most studied freshwater region, current reference libraries would not be able to detect 24-28% of species present or potentially invasive in the Great Lakes with a metabarcoding approach.
Key take-aways:
- eDNA metabarcoding that screens for both native and invasive species has significant potential, but gaps in reference libraries exist that limit implementation
- The authors identify 30 potentially or already established invasive species in the Great Lakes Basin that lack reference sequences
- Prioritizing sequencing of missing species for use in metabarcoding surveys in the Great Lakes biodiversity sampling would improve EDRR of invasive species with little extra effort
Management implications:
- This study demonstrates invasive species risk assessments can be used to prioritize invasive species missing from reference libraries, however improving coverage at a local and regional scale will likely fall to agencies tasked with managing invasive species
- Adding new reference sequences to libraries will improve coverage, but current approaches will unlikely achieve complete coverage without complementary approaches such as species-specific PCRs to help reduce false positives and missed detections
- Combining eDNA metabarcoding with conventional detection (e.g. traps, nets, electrofishing) would improve EDRR and calibration of fish community sampling